Wecomput provides professional antibody humanization service. For more details, please contract us.
Since the first monoclonal antibody muromonab (OKT3) was approved for clinical treatment, the development of therapeutic antibody has gone through several stages, including mouse antibody, chimeric antibody, humanized antibody and fully human antibody. In recent years, with the development of fully human phage display technology and humanized transgenic mice technology, fully human antibodies have been used more and more in clinic. However, humanized antibodies still account for more than 40% of the monoclonal antibodies in the market due to the technical accessibility and maturity, cost control, druggability and other reasons (Figure 1). Hybridoma monoclonal antibody with high affinity and physiological activity was obtained by immunizing mice, and its protein sequence was obtained by cloning the variable region gene of antibody, and then humanized antibody was obtained by humanization of antibody. It is still a routine strategy for the discovery of therapeutic antibodies. Since the modification of antibody humanization involves the change of amino acid sequence and even structure of antibody, the use of CADD technology for structure-based antibody humanization can not only retain or basically retain the affinity, specificity, physiological activity of mouse or chimeric antibody to antigen in the process of humanization, but also do not affect its drug developability (the expression level, physical and chemical properties, stability, etc).

Figure 1. Percentage of different humanized antibodies in marketed drugs (data updated to September 2018)
Sequence Numbering System for Variable Regions of Antibody, Definition of CDR and Basic Structure Features
Since the process of antibody humanization is accompanied by the analysis and modification of antibody sequence and structure, especially the variable region of heavy chain and light chain, it is necessary to understand the basic characteristics of antibody variable region sequence and structure before introducing the humanized transformation strategy. These characteristics are also the basis of antibody library construction, in vitro affinity maturation and other antibody engineering transformation.
The variable region gene of antibody is composed of immunoglobulin genes V, D, J fragments, which were were rearranged by VDJ (VJ rearrangement for light chain). The V gene encodes most of the region from the N-terminal of the variable region of the antibody to CDR3 (for light chain, V gene also encodes part of CDR3). CDR3 is usually co encoded by D gene and J gene (for light chain CDR3, V gene and J gene co encode), while J gene encodes the C-terminal of CDR3 and FR4. Further access to the Vbase database for specific coding sequences of human immunoglobulin VDJ germline genes (http://www2.mrc-lmb.cam.ac.uk/vbase/).
The antibody variable region (VH or VL, including the above VDJ gene components) is usually divided into framework region and complementary-determining region (CDR), in which the CDR is also known as hypervariable region. It is generally believed that CDR is involved in antigen-antibody binding, but the identification of CDR is determined by comparing a series of antibody sequences and comparing their variability. In addition, there are not only sequence differences but also length differences in the variable region of antibody, especially in the CDR. If the antibody is numbered in numerical order according to the conventional method, the number of the same conservative site in different antibodies is inconsistent. In order to overcome this problem, Kabat et al first invented the classical Kabat amino acid sequence numbering system and CDR definition system by comparing a large number of variable region sequences of antibodies. This system ensures the consistency of conservative amino acid sites in different antibodies by introducing letters such as a, b, c into the number of amino acid sites in the variable length region. Other commonly used similar CDR definitions and numbering systems include IMGT systems, Chothia systems, etc. Taking the variable heavy chain region of Pembrolizumab (Keytruda) antibody as an example, the CDR and amino acid number are as follows(figure 2):

Figure 2. Labeling of antibody CDR and amino acid sequence number
Structurally, the antibody variable region consists of nine β strands to form two β sheets, and each strand is connected by a loop. The positions of the three CDRs are generally corresponding to loop BC, loop C’C” and loop FG. (figure 3.). It is worth noting that, since CDR was first defined based on sequence variability as mentioned above, this loop structure in structural biology is not completely corresponding to the CDR, and the consistency between CDR and loop structure in different CDR definition systems is different.
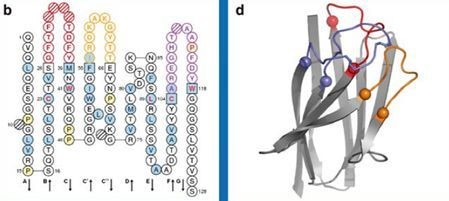
Figure 3. The structure of β strand in antibody variable region (AntibodyEngineering, Volume 2, Springer Protocols, 2010).
Further study on the structure of antigen-antibody complexes shows that only about one third of the amino acid residues in the CDR are directly involved in the binding with the antigen, and this part of the amino acid sequence changes more than the other parts of the CDR. This part of the amino acid residues are named as Specificity-Determine Residue (SDR), and the region containing these SDRs is named as abbreviated-CDRs (aCDRs). For Chothia CDR system, the relationship between hypervariable loop (HV), CDR and aCDR is shown below.(Figure 4.) :
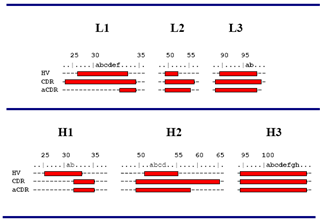
Figure 4. The relative positions of HV loop, CDR and ACDR (Chothia Numbering System)
Chothia et al. further found that the five CDRs except CDRH3 showed some fixed characteristics in sequence and structure, showing that they had some conservative sites in the CDR or Framework region relative to a specific CDR length. These sites were usually several fixed types of amino acid residues, which were important for maintaining the conformation of the CDR or loop region. These types of structures were called Canonical structure of antibody. In addition, Foote and Winter et al. found that there are some key amino acids supporting the CDR or loop region in the CDR adjacent region, including 16 variable regions of heavy chain and 14 variable regions of light chain, which are named Vernier Zone. Residues of Vernier Zone and Canonical Structure overlap to some extent. The locations of variable regions of heavy chain and light chain, CDR, aCDR, hypervariable loop, Canonical Structure Residues, Vernier Zone, etc. are shown below (Figure 5,6):

Figure 5. Sites of CDR and other structures in variable region of light chain
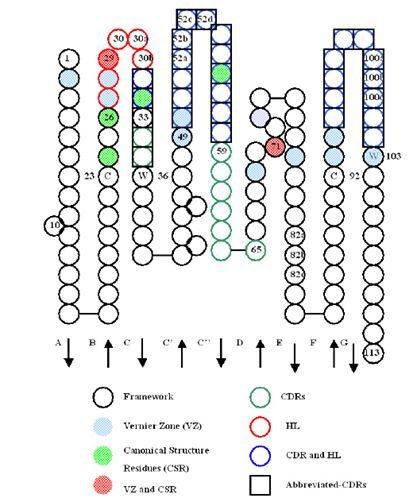
Figure 6. Sites of CDR and other structures in variable region of heavy chain
CDR Grafting, the most common and mature humanization method
As mentioned above, since the CDR is mainly involved in the interaction between antibody and antigen, the most conventional humanization idea is to graft these CDR that determine antigen-antibody interaction onto the human Framework region, thereby reducing the immunogenicity of mouse antibodies in the human body. The method of CDR grafting was first proposed in the late 1980s by Greg Winter team, the Nobel Prize winner of Chemistry Prize in 2018. After decades of development, the technology has been relatively mature and is also the most widely used and recognized humanization method in the industry.
The main steps of humanization method based on CDR grafting are as follows: 1) select the grafting region; 2) select the appropriate Framework donor (also called Framework receptor); 3) back mutation of key residues affecting the structure and function of antibodies.
– Select the Grafting Region
The ideal way before the antibody humanization was to obtain the crystal structure of the antibody-antigen complex, which helps to analyze which regions are involved in antigen-antibody interactions and thus select them as donors for CDR grafting. Unfortunately, such structural information usually does not exist in most cases. In this case, all six CDRs are usually used as donors for grafting. As mentioned earlier, since CDR was defined based on sequence, its structure overlaps with the loop region involved in antigen-antibody interaction, but it is not completely consistent, and the CDRs defined by different methods are also different, so that the region to be grafted can be selected according to the structure. For an antibody with known sequence and unknown structure, there are many commercial software (such as MOE, Schrodinger/Bioluminate, etc.) that can provide homology modeling of antibody structure.
If the humanization level or immunogenicity is taken into consideration, hoping to graft mouse antibody sequence as little as possible, the antibody can be humanized by grafting only the SDRs onto the human frameworks. Or prove that some CDRs may not be involved in the interaction of antigen and antibody by the mutant binding experiment, so only part of the CDR region is grafted. There are successful cases reports of these methods.
– Select Framework Donor (or Receptor)
In the development of CDR grafting technology, there are different methods for donor selection. Mouse CDRs were first directly grafted onto some human antibodies with known structures, and the homology of mouse antibodies and human antibodies in sequences was not considered. But the affinity of antibody obtained by this method is often significantly lower than that of mouse antibody or chimeric antibody. This makes researchers realize that different human antibody frameworks have different supporting effects on grafted CDR structures. So researchers consider human antibody Framework sequences with high homology with mouse antibodies as grafting donors, which is called Best Fit strategy. In addition to comparing the homology of the primary structure of the sequence, the consistency of the donor Framework with the grafted antibody on the Canonical structure can be considered when the donor Framework is selected, which is often helpful to maintain the CDR loop structure and the antigen-antibody affinity. Others consider the homology of CDR region instead of Framework, which is also based on the fact that the selected Framework sequence can better support the CDR structure.
There are two sources of Framework sequences. One is published human or humanized antibody sequences, which can be obtained through databases such as PDB and IMGT. However, these human sequences often have experienced high-frequency somatic mutations, and some mutations have been accumulated in the Framework region. These mutations may not be beneficial to candidate antibodies and have immunogenicity risks. The second source is immature human germline genes, which are less likely theoretically to cause immunogenicity without high frequency somatic mutation. In addition, studies have shown that the germline sequence as a Framework can provide better flexibility to the CDR region than the antibody of somatic cell maturation, so that the gafted CDR structure can be better maintained. At present, more and more germline gene framework rather than mature antibody framework is used as a framework donor for transplantation.
– Determine the Back Mutation of Residues
The CDR-grafting method often results in reduced affinity for the target antigen, which may be due to the change of some key sites supporting the structure of CDR in the process of humanization. In order to restore the affinity and physiological activity of the antibody, it is often necessary to carry out back mutation of the key residues of the CDR grafting antibody to retain its murine sequence.In this process, it is often necessary to consider the balance between affinity, function, developability and immunogenicity, humanization level. For different projects, the number of back mutations required is different, and the selection of mutation sites is also critical. As mentioned earlier, the residues in the Canonical Structure and Vernier Zone regions of the antibody may be important for maintaining the CDR loop conformations, and if mutations occur in these residues during the humanization, they may have an important impact on the CDR conformation and activity, so these key residues are usually given first consideration for back mutations. In addition, the residues within the Framework that are relatively close to the CDR region, such as those within 5A or 3A, may have intramolecular interactions with the CDR, or are directly involved in the interaction with antigens, which can also be included in the candidate sites of back mutations if mutated during humanization. By analyzing the known or modeled structure of antibody, combining the back mutations of heavy and light chain with different priority, and then conducting chessboard combination of heavy and light chains on these mutants with different back mutations, the drug candidates with less revertant mutations, and high affinity, physiological activity and developability are screened finally.
Humanization by Resurfacing Approach
Resurfacing is another common stragety for humanization of non-human antibodies, and its basis is similar to CDR grafting. But when selecting mutation sites, only the residues on the surface of antibody are taken into consideration during humanization, while the residues inside the antibody are maintained in the mouse antibody sequence. This greatly reduces the number of mutated amino acids required in the humanization process, which is beneficial for keeping the affinity for target antigen and physiological activity. The principle of this stragety is to consider that in the process of ADA production, B cells are required to recognize the B cell epitope on the surface of the antibody through BCR, and the internal mouse sequence is not recognized by B cells because it is not exposed, resulting in ADA reaction. However, due to the limited cases, the clinical safety and immunogenicity of resurfacing need to be further evaluated.
Screening of Humanized Candidate Antibodies using Framework Library
This technology combines CDR grafting with phage library technology. In some residue sites that are closely related to CDR such as Vernier Zone, randomized library construction is carried out. Using phage panning technology, humanized mutants with high affinity to target antigens are selected as drug candidates.
Antibody Humanization by Chain Reshuffling
This humanization method is also named as guided selection, which has been successfully used in the humanization process of TNFa antibody Humira. This method is also developed by Professor Greg Winter’s team. The idea is to fix the variable region VH of heavy chain of mouse antibody and construct a human shuffling library of light chain. The candidate molecules with high affinity to TNFa were selected by phage library technology, and then the VL sequence of this candidate molecule was selected to construct the human heavy chain shuffling library, and the drug candidate with full replacement of heavy and light chains into human antibodies were screened and selected.
Antibody Humanization Based on Human String Content (HSC) Scoring Algorithm
This method was first proposed by Lazar et al., and its point was that the linear T cell epitope that existed in murine antibody could combine with MHC II molecules on the surface of human antigen presenting cells and be recognized by TCR of T cell played a decisive role in the generation of ADA. In the conventional CDR grafting process, although most of the Framework region is replaced by human germline sequence, the potential T cell epitopes in the CDR region and the CDR-Framework junction region are ignored, resulting in immunogenicity of antibody. The scoring algorithm of HSC scores the T cell epitopes of the antibody. The higher the score is, the less the T cell epitopes are, and the higher the humanization is. The humanization process is to improve the HSC scoring by mutation, so as to reduce the immunogenicity.
Conclusions
The immunogenicity of antibodies has always been a hot issue in the industry. Although the causing of immunogenicity is not completely related to humanization level of antibodies, such as the subsequent development process and quality standards of antibody (such as aggregation), and the immune hyperstimulation status of patients with different indications will affect the generation of ADA in clinic, it is currently the consensus of the industry improve the humanization of antibodies as much as possible in the early development stage. How to choose humanization strategy has a very important influence on keeping the activity and drugability of antibodies. At present, CDR grafting is still the most common and widely accepted humanization strategy. Structure-based drug design technology can be used to design drugs rapidly and accurately, thus accelerating the development process of antibody drugs.
Reference
- Almagro J C, Fransson J. Humanization of antibodies[J]. Front Biosci, 2008, 1 3(1): 1619-1633.
- Safdari Y, Farajnia S, Asgharzadeh M, et al. Antibody humanization methods–a review and update[J]. Biotechnology and Genetic Engineering Reviews, 2013, 29(2): 175-186.
- Ahmadzadeh V, Farajnia S, Feizi M A H, et al. Antibody humanization methods for development of therapeutic applications[J]. Monoclonal antibodies in immunodiagnosis and immunotherapy, 2014, 33(2): 67-73.
- Lazar G A, Desjarlais J R, Jacinto J, et al. A molecular immunology approach to antibody humanization and functional optimization[J]. Molecular immunology, 2007, 44(8): 1986-1998.
- Antibody Engineering, Volume 1&2 (secondedition), edited by Roland Kontermannand Stefan Dübel, Springer Protocols(2010).